[ML] Classification with cilinical data: can we prevent heart failure through data analysis?
About data:
- This analysis includes heart failure clinical records dataset from Kaggle(https://www.kaggle.com/andrewmvd/heart-failure-clinical-data).
- The given CSV file contains such columns below.
- age: age of patients
- anaemia: decrease of red blood cells or hemoglobin (boolean: 0-normal, 1-anaemia)
- creatinine_phosphokinase: level of the CPK enzyme in the blood (mcg/L)
- diabetes: if the patient has diabetes (boolean: 0-normal, 1-diabetes)
- ejection_fraction: percentage of blood leaving the heart at each contraction (percentage: %)
- high_blood_pressure: If the patient has hypertension (boolean: 0-normal, 1-high blood pressure)
- platelets: platelets in the blood (kiloplatelets/mL)
- serum_creatinine: level of serum creatinine in the blood (mg/dL)
- serum_sodium: level of serum sodium in the blood (mEq/L)
- sex: sex of the patient (binary: 0-woman, 1-man)
- smoking: if the patient smokes or not (boolean: 0-no, 1-yes)
- time: follow-up period (days)
- DEATH_EVENT: If the patient deceased during the follow-up period (boolean: 0-alive, 1-dead)
Goals of the analysis
- Understanding the form of clinical data
- Applying pandas libary
- Acquring insights through data visualization
- Training models based on Scikit-learn
- Applying classification models and evaluating their performance
Step 1. Preparing dataset
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
1-1. Kaggle API setting
import os
os.environ['KAGGLE_USERNAME']="jisuleeoslo"
os.environ['KAGGLE_KEy']=""
1-2. Downloading data and unzipping data
!kaggle datasets download -d andrewmvd/heart-failure-clinical-data
heart-failure-clinical-data.zip: Skipping, found more recently modified local copy (use --force to force download)
import zipfile
with zipfile.ZipFile("heart-failure-clinical-data.zip","r") as zip_ref:
zip_ref.extractall()
1-3. Opening the csv file with Pandas library
df = pd.read_csv('heart_failure_clinical_records_dataset.csv')
ls
Step 2. Exploratory Data Analysis using descriptive statistics
2-1. Analyzing columns
df.head(5)
| age | anaemia | creatinine_phosphokinase | diabetes | ejection_fraction | high_blood_pressure | platelets | serum_creatinine | serum_sodium | sex | smoking | time | DEATH_EVENT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 75.0 | 0 | 582 | 0 | 20 | 1 | 265000.00 | 1.9 | 130 | 1 | 0 | 4 | 1 |
| 1 | 55.0 | 0 | 7861 | 0 | 38 | 0 | 263358.03 | 1.1 | 136 | 1 | 0 | 6 | 1 |
| 2 | 65.0 | 0 | 146 | 0 | 20 | 0 | 162000.00 | 1.3 | 129 | 1 | 1 | 7 | 1 |
| 3 | 50.0 | 1 | 111 | 0 | 20 | 0 | 210000.00 | 1.9 | 137 | 1 | 0 | 7 | 1 |
| 4 | 65.0 | 1 | 160 | 1 | 20 | 0 | 327000.00 | 2.7 | 116 | 0 | 0 | 8 | 1 |
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 299 entries, 0 to 298
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 299 non-null float64
1 anaemia 299 non-null int64
2 creatinine_phosphokinase 299 non-null int64
3 diabetes 299 non-null int64
4 ejection_fraction 299 non-null int64
5 high_blood_pressure 299 non-null int64
6 platelets 299 non-null float64
7 serum_creatinine 299 non-null float64
8 serum_sodium 299 non-null int64
9 sex 299 non-null int64
10 smoking 299 non-null int64
11 time 299 non-null int64
12 DEATH_EVENT 299 non-null int64
dtypes: float64(3), int64(10)
memory usage: 30.5 KB
df.describe()
| age | anaemia | creatinine_phosphokinase | diabetes | ejection_fraction | high_blood_pressure | platelets | serum_creatinine | serum_sodium | sex | smoking | time | DEATH_EVENT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 299.000000 | 299.000000 | 299.000000 | 299.000000 | 299.000000 | 299.000000 | 299.000000 | 299.00000 | 299.000000 | 299.000000 | 299.00000 | 299.000000 | 299.00000 |
| mean | 60.833893 | 0.431438 | 581.839465 | 0.418060 | 38.083612 | 0.351171 | 263358.029264 | 1.39388 | 136.625418 | 0.648829 | 0.32107 | 130.260870 | 0.32107 |
| std | 11.894809 | 0.496107 | 970.287881 | 0.494067 | 11.834841 | 0.478136 | 97804.236869 | 1.03451 | 4.412477 | 0.478136 | 0.46767 | 77.614208 | 0.46767 |
| min | 40.000000 | 0.000000 | 23.000000 | 0.000000 | 14.000000 | 0.000000 | 25100.000000 | 0.50000 | 113.000000 | 0.000000 | 0.00000 | 4.000000 | 0.00000 |
| 25% | 51.000000 | 0.000000 | 116.500000 | 0.000000 | 30.000000 | 0.000000 | 212500.000000 | 0.90000 | 134.000000 | 0.000000 | 0.00000 | 73.000000 | 0.00000 |
| 50% | 60.000000 | 0.000000 | 250.000000 | 0.000000 | 38.000000 | 0.000000 | 262000.000000 | 1.10000 | 137.000000 | 1.000000 | 0.00000 | 115.000000 | 0.00000 |
| 75% | 70.000000 | 1.000000 | 582.000000 | 1.000000 | 45.000000 | 1.000000 | 303500.000000 | 1.40000 | 140.000000 | 1.000000 | 1.00000 | 203.000000 | 1.00000 |
| max | 95.000000 | 1.000000 | 7861.000000 | 1.000000 | 80.000000 | 1.000000 | 850000.000000 | 9.40000 | 148.000000 | 1.000000 | 1.00000 | 285.000000 | 1.00000 |
Applying df.describe(), one can check:
- Wheter binary data are balanced or imblanced. (For example, the mean of sex is 0.64 and that of smoking is 0.32. This implies there are more data of patients who are men than women. The data include more of non-smokers than smokers.)
- Whether there are extreme outliers
- How is the increase rate of each percentile
- Whether some variables are correlated
2-2. Drawing histogram on numerical data
import seaborn as sns
sns.histplot(data=df, x='age')
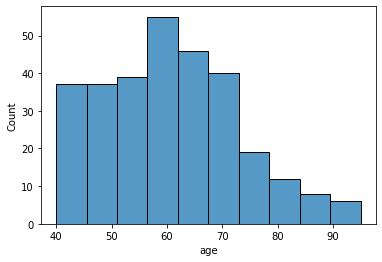
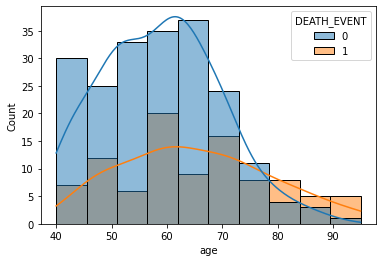
sns.histplot(data=df, x='age', hue='DEATH_EVENT', kde=True)
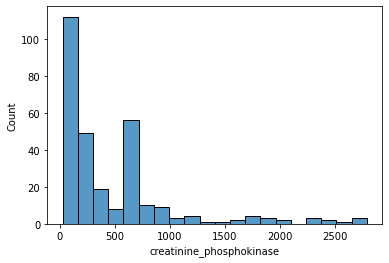
sns.histplot(data=df.loc[df['creatinine_phosphokinase'] < 3000, 'creatinine_phosphokinase'])
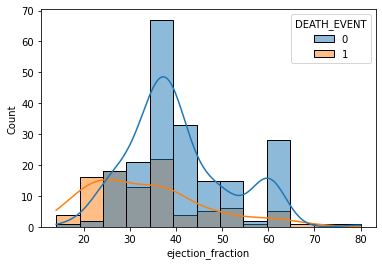
sns.histplot(data=df, x='ejection_fraction', bins=13, hue='DEATH_EVENT', kde=True)
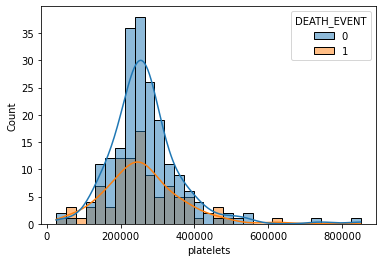
sns.histplot(data=df, x='platelets', hue='DEATH_EVENT', kde=True)
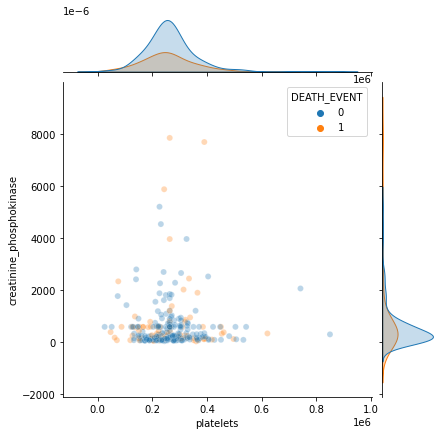
sns.jointplot(x='platelets', y='creatinine_phosphokinase', hue='DEATH_EVENT', data=df, alpha=0.3)
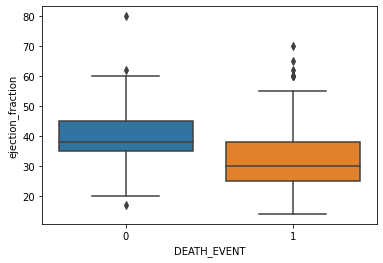
2-3. Drawing boxplot on categorical data
sns.boxplot(x='DEATH_EVENT', y='ejection_fraction', data=df)
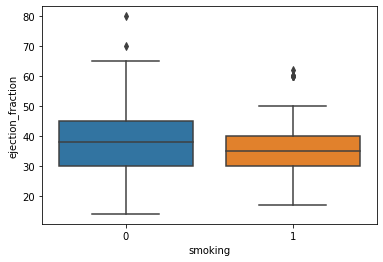
sns.boxplot(x='smoking', y='ejection_fraction', data=df)
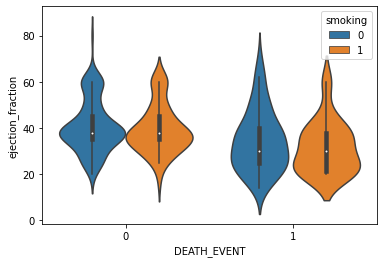
sns.violinplot(x='DEATH_EVENT', y='ejection_fraction', hue='smoking', data=df)
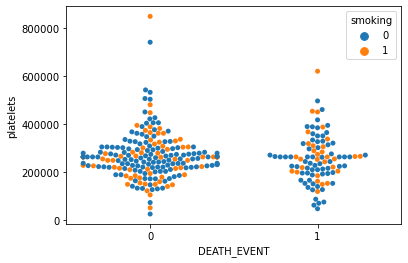
sns.swarmplot(x='DEATH_EVENT', y='platelets', hue='smoking', data=df)
Step 3. Data pre-processing for modeling
3-1. Feature scaling continous data with StandardScaler
from sklearn.preprocessing import StandardScaler
df.head(5)
| age | anaemia | creatinine_phosphokinase | diabetes | ejection_fraction | high_blood_pressure | platelets | serum_creatinine | serum_sodium | sex | smoking | time | DEATH_EVENT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 75.0 | 0 | 582 | 0 | 20 | 1 | 265000.00 | 1.9 | 130 | 1 | 0 | 4 | 1 |
| 1 | 55.0 | 0 | 7861 | 0 | 38 | 0 | 263358.03 | 1.1 | 136 | 1 | 0 | 6 | 1 |
| 2 | 65.0 | 0 | 146 | 0 | 20 | 0 | 162000.00 | 1.3 | 129 | 1 | 1 | 7 | 1 |
| 3 | 50.0 | 1 | 111 | 0 | 20 | 0 | 210000.00 | 1.9 | 137 | 1 | 0 | 7 | 1 |
| 4 | 65.0 | 1 | 160 | 1 | 20 | 0 | 327000.00 | 2.7 | 116 | 0 | 0 | 8 | 1 |
# dividing data into numerical, categorical and output
X_num= df[['age', 'creatinine_phosphokinase', 'ejection_fraction', 'platelets',
'serum_creatinine', 'serum_sodium', 'time']]
X_cat = df[['anaemia', 'diabetes', 'high_blood_pressure', 'sex', 'smoking']]
y = df['DEATH_EVENT']
# Feature scaling continous data and integrating them into numpy array
scaler = StandardScaler()
scaler.fit(X_num)
X_scaled = scaler.transform(X_num)
X_scaled
array([[ 1.19294523e+00, 1.65728387e-04, -1.53055953e+00, ...,
4.90056987e-01, -1.50403612e+00, -1.62950241e+00],
[-4.91279276e-01, 7.51463953e+00, -7.07675018e-03, ...,
-2.84552352e-01, -1.41976151e-01, -1.60369074e+00],
[ 3.50832977e-01, -4.49938761e-01, -1.53055953e+00, ...,
-9.09000174e-02, -1.73104612e+00, -1.59078490e+00],
...,
[-1.33339153e+00, 1.52597865e+00, 1.85495776e+00, ...,
-5.75030855e-01, 3.12043840e-01, 1.90669738e+00],
[-1.33339153e+00, 1.89039811e+00, -7.07675018e-03, ...,
5.92615005e-03, 7.66063830e-01, 1.93250906e+00],
[-9.12335403e-01, -3.98321274e-01, 5.85388775e-01, ...,
1.99578485e-01, -1.41976151e-01, 1.99703825e+00]])
X_scaled = pd.DataFrame(data=X_scaled, index=X_num.index, columns=X_num.columns)
X = pd.concat([X_scaled, X_cat], axis=1)
X
| age | creatinine_phosphokinase | ejection_fraction | platelets | serum_creatinine | serum_sodium | time | anaemia | diabetes | high_blood_pressure | sex | smoking | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.192945 | 0.000166 | -1.530560 | 1.681648e-02 | 0.490057 | -1.504036 | -1.629502 | 0 | 0 | 1 | 1 | 0 |
| 1 | -0.491279 | 7.514640 | -0.007077 | 7.535660e-09 | -0.284552 | -0.141976 | -1.603691 | 0 | 0 | 0 | 1 | 0 |
| 2 | 0.350833 | -0.449939 | -1.530560 | -1.038073e+00 | -0.090900 | -1.731046 | -1.590785 | 0 | 0 | 0 | 1 | 1 |
| 3 | -0.912335 | -0.486071 | -1.530560 | -5.464741e-01 | 0.490057 | 0.085034 | -1.590785 | 1 | 0 | 0 | 1 | 0 |
| 4 | 0.350833 | -0.435486 | -1.530560 | 6.517986e-01 | 1.264666 | -4.682176 | -1.577879 | 1 | 1 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 294 | 0.098199 | -0.537688 | -0.007077 | -1.109765e+00 | -0.284552 | 1.447094 | 1.803451 | 0 | 1 | 1 | 1 | 1 |
| 295 | -0.491279 | 1.278215 | -0.007077 | 6.802472e-02 | -0.187726 | 0.539054 | 1.816357 | 0 | 0 | 0 | 0 | 0 |
| 296 | -1.333392 | 1.525979 | 1.854958 | 4.902082e+00 | -0.575031 | 0.312044 | 1.906697 | 0 | 1 | 0 | 0 | 0 |
| 297 | -1.333392 | 1.890398 | -0.007077 | -1.263389e+00 | 0.005926 | 0.766064 | 1.932509 | 0 | 0 | 0 | 1 | 1 |
| 298 | -0.912335 | -0.398321 | 0.585389 | 1.348231e+00 | 0.199578 | -0.141976 | 1.997038 | 0 | 0 | 0 | 1 | 1 |
299 rows × 12 columns
X.head()
| age | creatinine_phosphokinase | ejection_fraction | platelets | serum_creatinine | serum_sodium | time | anaemia | diabetes | high_blood_pressure | sex | smoking | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.192945 | 0.000166 | -1.530560 | 1.681648e-02 | 0.490057 | -1.504036 | -1.629502 | 0 | 0 | 1 | 1 | 0 |
| 1 | -0.491279 | 7.514640 | -0.007077 | 7.535660e-09 | -0.284552 | -0.141976 | -1.603691 | 0 | 0 | 0 | 1 | 0 |
| 2 | 0.350833 | -0.449939 | -1.530560 | -1.038073e+00 | -0.090900 | -1.731046 | -1.590785 | 0 | 0 | 0 | 1 | 1 |
| 3 | -0.912335 | -0.486071 | -1.530560 | -5.464741e-01 | 0.490057 | 0.085034 | -1.590785 | 1 | 0 | 0 | 1 | 0 |
| 4 | 0.350833 | -0.435486 | -1.530560 | 6.517986e-01 | 1.264666 | -4.682176 | -1.577879 | 1 | 1 | 0 | 0 | 0 |
3-2. Dividing the data into train and test dataset
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
X_train
| age | creatinine_phosphokinase | ejection_fraction | platelets | serum_creatinine | serum_sodium | time | anaemia | diabetes | high_blood_pressure | sex | smoking | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 79 | -0.491279 | -0.253792 | 0.585389 | 0.621074 | -0.478205 | 0.766064 | -0.726094 | 0 | 0 | 1 | 0 | 0 |
| 282 | -1.586025 | -0.534591 | -0.684180 | -0.495266 | 2.329754 | -1.958056 | 1.545334 | 0 | 0 | 0 | 1 | 1 |
| 297 | -1.333392 | 1.890398 | -0.007077 | -1.263389 | 0.005926 | 0.766064 | 1.932509 | 0 | 0 | 0 | 1 | 1 |
| 233 | -0.659702 | 0.129209 | -0.007077 | 0.682524 | 0.005926 | 0.085034 | 1.016195 | 1 | 0 | 0 | 1 | 1 |
| 106 | -0.491279 | 0.171536 | 0.585389 | -0.003667 | -0.090900 | 0.085034 | -0.545412 | 0 | 0 | 0 | 1 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 203 | -0.070223 | -0.539753 | -1.107370 | -0.525991 | 2.039276 | -0.141976 | 0.732266 | 0 | 0 | 1 | 1 | 1 |
| 255 | -0.743913 | -0.403483 | -0.684180 | 0.723490 | -0.381379 | 1.220084 | 1.106535 | 1 | 1 | 1 | 1 | 1 |
| 72 | 2.035057 | 5.471619 | -0.260991 | -0.208500 | -0.381379 | -1.050016 | -0.751905 | 0 | 0 | 0 | 1 | 1 |
| 235 | 1.361368 | -0.488136 | 1.008578 | 1.460889 | -0.284552 | 0.085034 | 1.016195 | 1 | 0 | 1 | 1 | 0 |
| 37 | 1.782424 | 0.281997 | 1.008578 | 0.590349 | -0.381379 | 1.901114 | -1.293951 | 1 | 1 | 1 | 0 | 0 |
200 rows × 12 columns
Step 4. Applying classification models
4-1. Applying Logistic Regression model
from sklearn.linear_model import LogisticRegression
model_lr = LogisticRegression()
model_lr.fit(X_train, y_train)
LogisticRegression()
4-2. Evaluating classification performance of Logistic Regression model
from sklearn.metrics import classification_report
pred = model_lr.predict(X_test)
print(classification_report(y_test, pred))
precision recall f1-score support
0 0.86 0.90 0.88 70
1 0.73 0.66 0.69 29
accuracy 0.83 99
macro avg 0.80 0.78 0.79 99
weighted avg 0.82 0.83 0.83 99
4-3. Applying XGBoost model
from xgboost import XGBClassifier
model_xgb = XGBClassifier()
model_xgb.fit(X_train, y_train)
XGBClassifier(base_score=0.5, booster='gbtree', colsample_bylevel=1,
colsample_bynode=1, colsample_bytree=1, gamma=0, gpu_id=-1,
importance_type='gain', interaction_constraints='',
learning_rate=0.300000012, max_delta_step=0, max_depth=6,
min_child_weight=1, missing=nan, monotone_constraints='()',
n_estimators=100, n_jobs=8, num_parallel_tree=1, random_state=0,
reg_alpha=0, reg_lambda=1, scale_pos_weight=1, subsample=1,
tree_method='exact', validate_parameters=1, verbosity=None)
4-4. Evaluating classification performance of XGBoost model
pred = model_xgb.predict(X_test)
print(classification_report(y_test, pred))
precision recall f1-score support
0 0.90 0.91 0.91 70
1 0.79 0.76 0.77 29
accuracy 0.87 99
macro avg 0.84 0.84 0.84 99
weighted avg 0.87 0.87 0.87 99
Too good to be true!
- The accuracy is almost 90%. Maybe there is something missing!
4-5. Finding important features
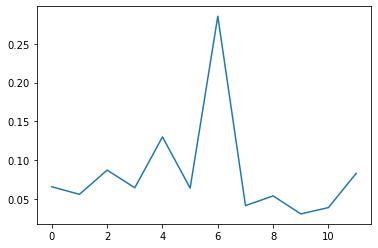
# What is the important features of XGBClassifier model?
plt.plot(model_xgb.feature_importances_)
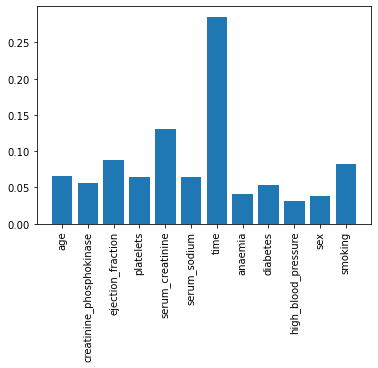
plt.bar(X.columns, model_xgb.feature_importances_)
plt.xticks(rotation=90)
plt.show()
sns.histplot(x='time', data=df, hue='DEATH_EVENT', kde=True)
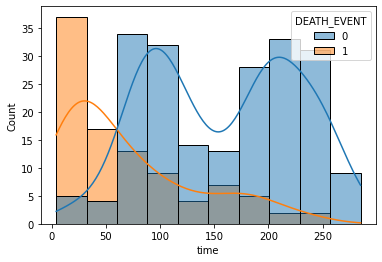
Time as the most important feature?
- Time refers to follow-up period.
- Time strongly depends on the death event.
- Therefore, It could have been better to get rid of ‘time’data.
4-6. Training again without time data
X_num= df[['age', 'creatinine_phosphokinase', 'ejection_fraction', 'platelets',
'serum_creatinine', 'serum_sodium']]
X_cat = df[['anaemia', 'diabetes', 'high_blood_pressure', 'sex', 'smoking']]
y = df['DEATH_EVENT']
scaler = StandardScaler()
scaler.fit(X_num)
X_scaled = scaler.transform(X_num)
X_scaled = pd.DataFrame(data=X_scaled, index=X_num.index, columns=X_num.columns)
X = pd.concat([X_scaled, X_cat], axis=1)
X.head()
| age | creatinine_phosphokinase | ejection_fraction | platelets | serum_creatinine | serum_sodium | anaemia | diabetes | high_blood_pressure | sex | smoking | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.192945 | 0.000166 | -1.530560 | 1.681648e-02 | 0.490057 | -1.504036 | 0 | 0 | 1 | 1 | 0 |
| 1 | -0.491279 | 7.514640 | -0.007077 | 7.535660e-09 | -0.284552 | -0.141976 | 0 | 0 | 0 | 1 | 0 |
| 2 | 0.350833 | -0.449939 | -1.530560 | -1.038073e+00 | -0.090900 | -1.731046 | 0 | 0 | 0 | 1 | 1 |
| 3 | -0.912335 | -0.486071 | -1.530560 | -5.464741e-01 | 0.490057 | 0.085034 | 1 | 0 | 0 | 1 | 0 |
| 4 | 0.350833 | -0.435486 | -1.530560 | 6.517986e-01 | 1.264666 | -4.682176 | 1 | 1 | 0 | 0 | 0 |
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1)
model_lr = LogisticRegression(max_iter=1000)
model_lr.fit(X_train, y_train)
LogisticRegression(max_iter=1000)
pred = model_lr.predict(X_test)
print(classification_report(y_test, pred))
precision recall f1-score support
0 0.78 0.92 0.84 64
1 0.64 0.35 0.45 26
accuracy 0.76 90
macro avg 0.71 0.63 0.65 90
weighted avg 0.74 0.76 0.73 90
model_xgb = XGBClassifier()
model_xgb.fit(X_train, y_train)
XGBClassifier(base_score=0.5, booster='gbtree', colsample_bylevel=1,
colsample_bynode=1, colsample_bytree=1, gamma=0, gpu_id=-1,
importance_type='gain', interaction_constraints='',
learning_rate=0.300000012, max_delta_step=0, max_depth=6,
min_child_weight=1, missing=nan, monotone_constraints='()',
n_estimators=100, n_jobs=8, num_parallel_tree=1, random_state=0,
reg_alpha=0, reg_lambda=1, scale_pos_weight=1, subsample=1,
tree_method='exact', validate_parameters=1, verbosity=None)
pred = model_xgb.predict(X_test)
print(classification_report(y_test, pred))
precision recall f1-score support
0 0.81 0.88 0.84 64
1 0.62 0.50 0.55 26
accuracy 0.77 90
macro avg 0.72 0.69 0.70 90
weighted avg 0.76 0.77 0.76 90
plt.bar(X.columns, model_xgb.feature_importances_)
plt.xticks(rotation=90)
plt.show()
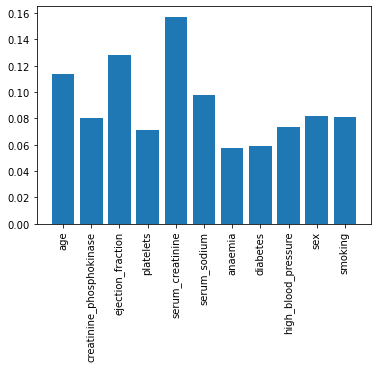
4-7. What is the relationship between the two most important features?
- Serum_creatinine as the most important feature.
- Ejection_fraction as the second important feature.
sns.jointplot(x='ejection_fraction', y='serum_creatinine',hue='DEATH_EVENT',data=df)
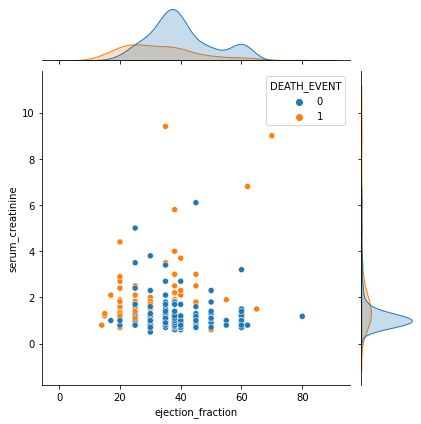
Step 5. In-depth analysis on the learning results
5-1. Idetifying Precision-Recall curve
from sklearn.metrics import plot_precision_recall_curve
plot_precision_recall_curve(model_lr, X_test, y_test)
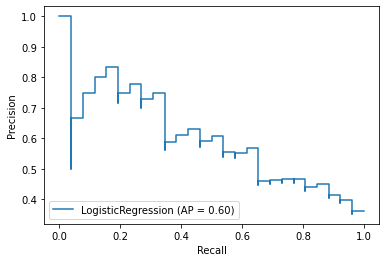
plot_precision_recall_curve(model_xgb, X_test, y_test)
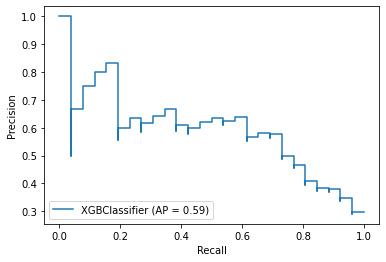
fig = plt.figure()
ax = fig.gca()
plot_precision_recall_curve(model_lr, X_test, y_test,ax=ax)
plot_precision_recall_curve(model_xgb, X_test, y_test,ax=ax)
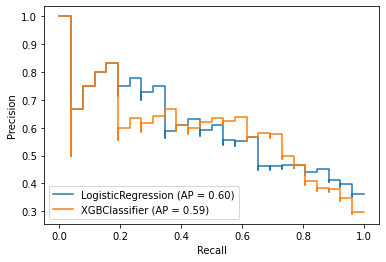
P*R curve implies XGBClassifier yielded better performance on the data
- The closer to 1, the better performance.
5-2. Identifying ROC curve
from sklearn.metrics import plot_roc_curve
fig = plt.figure()
ax = fig.gca()
plot_roc_curve(model_lr, X_test, y_test,ax=ax)
plot_roc_curve(model_xgb, X_test, y_test,ax=ax)
plt.show()
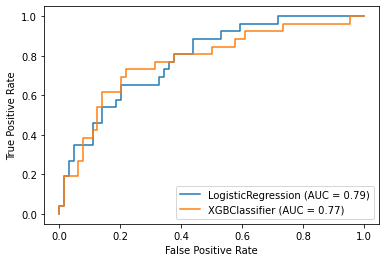
ROC curve implies LogisticRegression yielded better performance on the data
- The faster reaching 1 while keeping low FPS, the better performance.
- The higher Area Under Cover(AUC), the better performance.
- In result, it is difficult to say which model is better than the other.
